Modern day plant breeding and plant research is based on the understanding of plant genetics.
The advance in Next Generation Sequencing technologies NGS and their availability enabled more and more crop genes and even whole genomes being discovered, deciphered, and better understood.
Genome Wide Association Studies GWAS, Quantitative Trait Loci QTL mapping and other plant genetics research studies, are now possible and common.
Further pre-breeding and Marker Assisted Selection MAS breeding, requires screening and genotyping of multiple samples for multiple genes/markers.
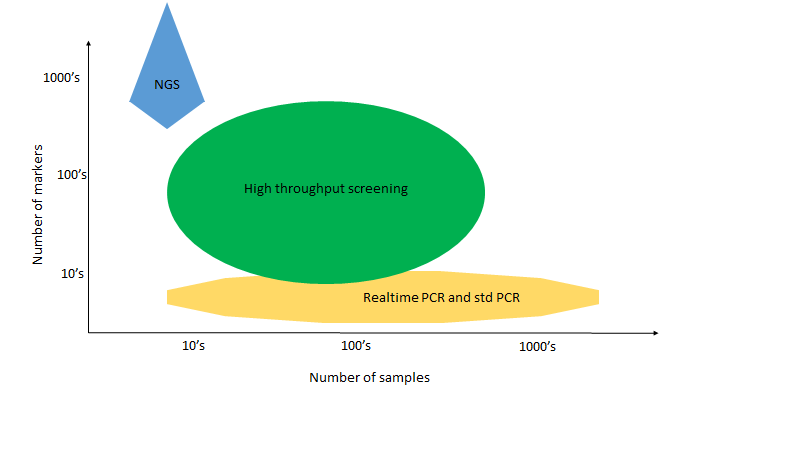
High throughput genotyping of 100’s to 1000’s of plants on 10’s and even hundreds of genes is a totally different task from analyzing whole genomes for a few samples or from genotyping several genes on thousands of samples.
In this post we will focus on different molecular methods to perform high throughput genotyping on 10’s to 100’s of DNA markers.
DNA Genotyping data is often described and calculated by the term DATA POINT (dp). Data point is the test result for 1 sample with 1 marker.
For example, when you are tested for the presence of covid19 the test result is 1dp = 1 person X 1(covid19) test.
DNA marker screening or GenoTyping is a matrix of the sample size: plant population screening size, and the DNA markers range/size. For different genetics studies or even marker assisted breeding strategies, different kinds of genetics screening are required.
Sometimes genotyping of many samples with few DNA markers, and sometimes on many more markers.
NGS tools are designated for analyzing infinite DNA/RNA sequences on limited samples from a few to 10’s or as many as hundreds.
Other simpler PCR and now Realtime PCR genotyping methods are best for Genotyping 100’s to 1000’s and even more samples, on a limited number of DNA markers.
For other applications there is a need for genotyping of 10’s to 100’s of markers on 100’s and even 1000’s of samples.
We will now focus on technologies making these screenings possible, practical and affordable.
GBS will not be mentioned or other NGS based methods. This will be addressed in another post.
Microarrays are not mentioned either, as they are too, designated to analyze >1000’s of markers on few to several samples.
This technology uses 3 primers per SNP. Works only on SNP’s not Indels.
https://www.illumina.com/Documents/
A 1536 SNPs project for 96 to 384 samples will take some 3 days which is very impressive.
There are many papers from the last years that made use of this platform.
The service cost for 96 samples X 1536 markers is somewhere on the lines of 200+ USD per sample.
While searching for the best tools for our needs, the added cost of the instrumentation and service, together with the Illumina kits and the work required for working with this platform, made it out of budget reach for us.
A microfluidics PCR platform enabling a matrix of 96 samples on 96 markers.
The Realtime PCR can work with Taqman or KASP. https://www.gene-g.com/what-is-allelic-discrimination-genotyping/
Requires special microfluidics cells; each cell of 96 markers X96 samples is non-reusable and the cost is about a few hundreds of $’s.
For an array of 96 markers, you also need to add the cost for primers and probes (if Taqman) and real-time PCR mixes.
A single new SNP design and order cost even in minimal amounts is ~280euro (for Taqman), So a 96 samples X96 markers new array of SNPs will cost you 280euroX96 =~ >25K Euro.
If you are doing KASP then this could be somewhat reduced, But we found that KASP will not work on every SNP and is problematic on InDels, and somehow more difficult all around.
A fully automated system based on Douglas Scientific Array Tape PCR.
Allows a PCR in an 800nl volume making it highly economic.
I have no personal experience with this system.
The advantage as I see it is that it is not a multiplex PCR method, but rather a method to minimize the singleplex reaction, and thus to reduce costs.
Technically, this can be an advantage.
Works with KASP and Taqman.
https://www.agenabio.com/products/massarray-system/
The technology is based on PCR followed by a primer extension reaction.
Then all is transferred through automated liquid handling into a chip and analyzed through a Mass spectrometer (MALDI TOF).
SNP, short InDels are automatically analyzed by the software and exported easily to excel.
We use the designated primers/assay design tool and the success rate of SNPs and short InDel assays is nearly 100%.
The design tool is actually one of the best primers design tools we ever worked with, and it enables a 60 plex (!!) of different SNPs, primers design.
We actually ran more than a few of these 60plex assays.
Requires special Agena Chips which are not cheap, but the chips kits contain all reagents required for the run including enzymes and everything.
We receive service from the European Agena branch and they are prompt and efficient and highly professional.
Most of our problems were solved remotely, even before the pandemic.
We selected this system because of the flexibility and the cost/dp and the simplicity of operating it.
We use it both for routine genotyping of same multiplexes: trait genes or polymorphic fingerprint SNPs for various crops.
And we also, many times, run newly designed onetime multiplexes for several applications such as: Gene-Trait discovery, and SNPs selection for NGS/GBS data, and other applications.
Screening costs can be <0.2USD/dp, hence a project of 384 samples on 180 SNPs is a total dp of: 69,120.
This project of 384 X 180 SNPs, In the Agena format, is a 3 chip project, hence <3 days of TAT (turnaround time) for results, and the screening cost is <13K$ for full service.
https://www.gene-g.com/services/ (you can try use our cost calculator under General Genotyping for cost estimation)
This is an intermediate between NGS methods and high throughput screening methods. Targeted NGS enables screening >100 and up to several thousands of genes, on hundreds of samples.
There are several companies that now offer a wide screening of genes/markers through targeted Next Generation Sequencing. Together with our customers, we have some good experience with 2 service providers:
https://igatechnology.com/agrigenomics-solutions/
https://rapid-genomics.com/applications/agriculture/capture-seq/
These methods require access to NGS instrumentation therefore it is mainly offered as full service.
The great advantage of these methods is that the output is DNA sequences. Opposed to all above mentioned where you get a SNP or an InDel call, with targeted NGS you can actually read a DNA sequence for each sample in the targeted region. This makes the analysis of the data more challenging yet it can serve better as a sequencing tool rather than just a simple SNP/Indel typing tool.
Costs for these tests range from 25$ for a 100 SNPs to 30 euro for 5000 SNPs+ 100bp around the SNP.
In the past 2 decades DNA sequencing and genotyping technologies are continuously developing and evolving.
This is very useful for the industry and allows for projects once thought almost impossible to have accurate results in a short time span.
This constant change and development make it also a great challenge in selecting the “technology of choice” for your own lab, or for a certain project. It’s somehow like buying a computer in the 80’s or 90’s when you were almost sure that in a few months your new computer will already be outdated. Nevertheless, all the above-mentioned methods have advantages.
The main idea is to always match between the requirements of the project and your limitations: costs, time and select the best method to achieve your tasks.
For over 6 years we at Gene-G provide full service high throughput genotyping, and ready-to-use-kits and we are also constantly involved in genes/markers discovery projects.
We would be more than happy to consult.

Dagan Mor, Gene-G
visibility_offDisable flashes
titleMark headings
settingsBackground Color
zoom_outZoom out
zoom_inZoom in
remove_circle_outlineDecrease font
add_circle_outlineIncrease font
spellcheckReadable font
brightness_highBright contrast
brightness_lowDark contrast
format_underlinedUnderline links
font_downloadMark links